Iteration
Jeff Stevens
2025-03-26
Introduction
The problems
- Run the same model on multiple data sets
- Create the same plot multiple data sets
- Read in data files from multiple subjects
Set-up
Iteration
For loops
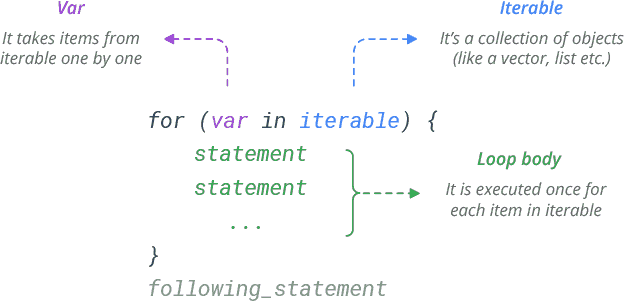
For loops
for (counter in min:max) {
# What you want repeated.
# Index counter-specific vector with [counter]
}Building objects with for loops
for (i in 1:nrow(penguins)) {
bill_size[i] <- penguins$bill_length_mm[i] * penguins$bill_depth_mm[i]
}Error: object 'bill_size' not foundFor loops with vectors
month.name [1] "January" "February" "March" "April" "May" "June"
[7] "July" "August" "September" "October" "November" "December" for (i in month.name) {
print(paste0(i, " has ", str_length(i), " letters."))
}[1] "January has 7 letters."
[1] "February has 8 letters."
[1] "March has 5 letters."
[1] "April has 5 letters."
[1] "May has 3 letters."
[1] "June has 4 letters."
[1] "July has 4 letters."
[1] "August has 6 letters."
[1] "September has 9 letters."
[1] "October has 7 letters."
[1] "November has 8 letters."
[1] "December has 8 letters."R is vectorized
Most R functions apply to vectors, so no need for for loops
bill_size <- penguins$bill_length_mm * penguins$bill_depth_mm
head(bill_size)[1] 731.17 687.30 725.40 NA 708.31 809.58print(paste0(month.name, " has ", str_length(month.name), " letters.")) [1] "January has 7 letters." "February has 8 letters."
[3] "March has 5 letters." "April has 5 letters."
[5] "May has 3 letters." "June has 4 letters."
[7] "July has 4 letters." "August has 6 letters."
[9] "September has 9 letters." "October has 7 letters."
[11] "November has 8 letters." "December has 8 letters." Lagged and leading values
To compute based on previous or subsequent values, use dplyr::lag() and dplyr::lead()
data.frame(trial = 1:8, cond = rep(1:2, each = 4), resp = c(1, 3, 4, 6, 1, 4, 8, 10)) |>
mutate(behind = lag(trial),
ahead = lead(trial),
diff = resp - lag(resp)) trial cond resp behind ahead diff
1 1 1 1 NA 2 NA
2 2 1 3 1 3 2
3 3 1 4 2 4 1
4 4 1 6 3 5 2
5 5 2 1 4 6 -5
6 6 2 4 5 7 3
7 7 2 8 6 8 4
8 8 2 10 7 NA 2Lagged and leading values
To compute based on previous or subsequent values, use dplyr::lag() and dplyr::lead()
data.frame(trial = 1:8, cond = rep(1:2, each = 4), resp = c(1, 3, 4, 6, 1, 4, 8, 10)) |>
mutate(behind = lag(trial),
ahead = lead(trial),
diff = resp - lag(resp),
diff = ifelse(cond != lag(cond), NA, diff)) trial cond resp behind ahead diff
1 1 1 1 NA 2 NA
2 2 1 3 1 3 2
3 3 1 4 2 4 1
4 4 1 6 3 5 2
5 5 2 1 4 6 NA
6 6 2 4 5 7 3
7 7 2 8 6 8 4
8 8 2 10 7 NA 2Numbering groups
Renumber row numbers based on groups
data.frame(trial = 1:8, cond = rep(1:2, each = 4)) |>
mutate(.by = cond,
cond_trial = row_number()) trial cond cond_trial
1 1 1 1
2 2 1 2
3 3 1 3
4 4 1 4
5 5 2 1
6 6 2 2
7 7 2 3
8 8 2 4Mapping over multiple arguments
Mapping with {purrr}
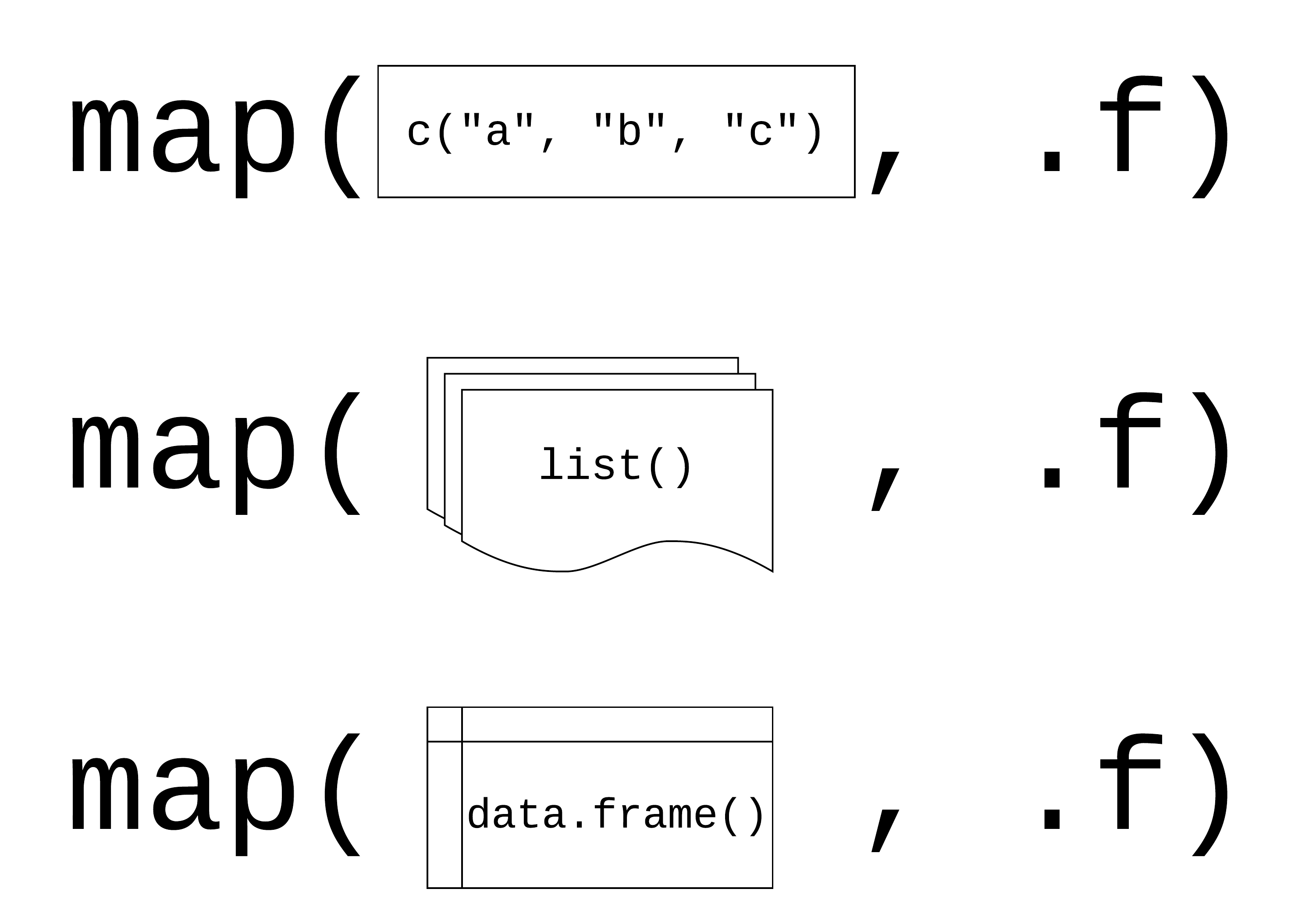

Mapping functions
purrr::map() applies functions repeatedly across data
$bill_length_mm
[1] 43.92193
$bill_depth_mm
[1] 17.15117
$flipper_length_mm
[1] 200.9152What kind of data type does map() return?
Mapping function
Need different data types as output?
Mapping function
Use split() like dplyr::group_by()
penguins |>
split(penguins$species) |> # split() inputs vector (not column)
map(~ lm(bill_length_mm ~ bill_depth_mm, data = .x))$Adelie
Call:
lm(formula = bill_length_mm ~ bill_depth_mm, data = .x)
Coefficients:
(Intercept) bill_depth_mm
23.068 0.857
$Chinstrap
Call:
lm(formula = bill_length_mm ~ bill_depth_mm, data = .x)
Coefficients:
(Intercept) bill_depth_mm
13.428 1.922
$Gentoo
Call:
lm(formula = bill_length_mm ~ bill_depth_mm, data = .x)
Coefficients:
(Intercept) bill_depth_mm
17.230 2.021 Mapping function
Create multiple plots
penguins |>
split(penguins$species) |>
map(~ ggplot(.x, aes(bill_length_mm, bill_depth_mm)) + geom_point())$Adelie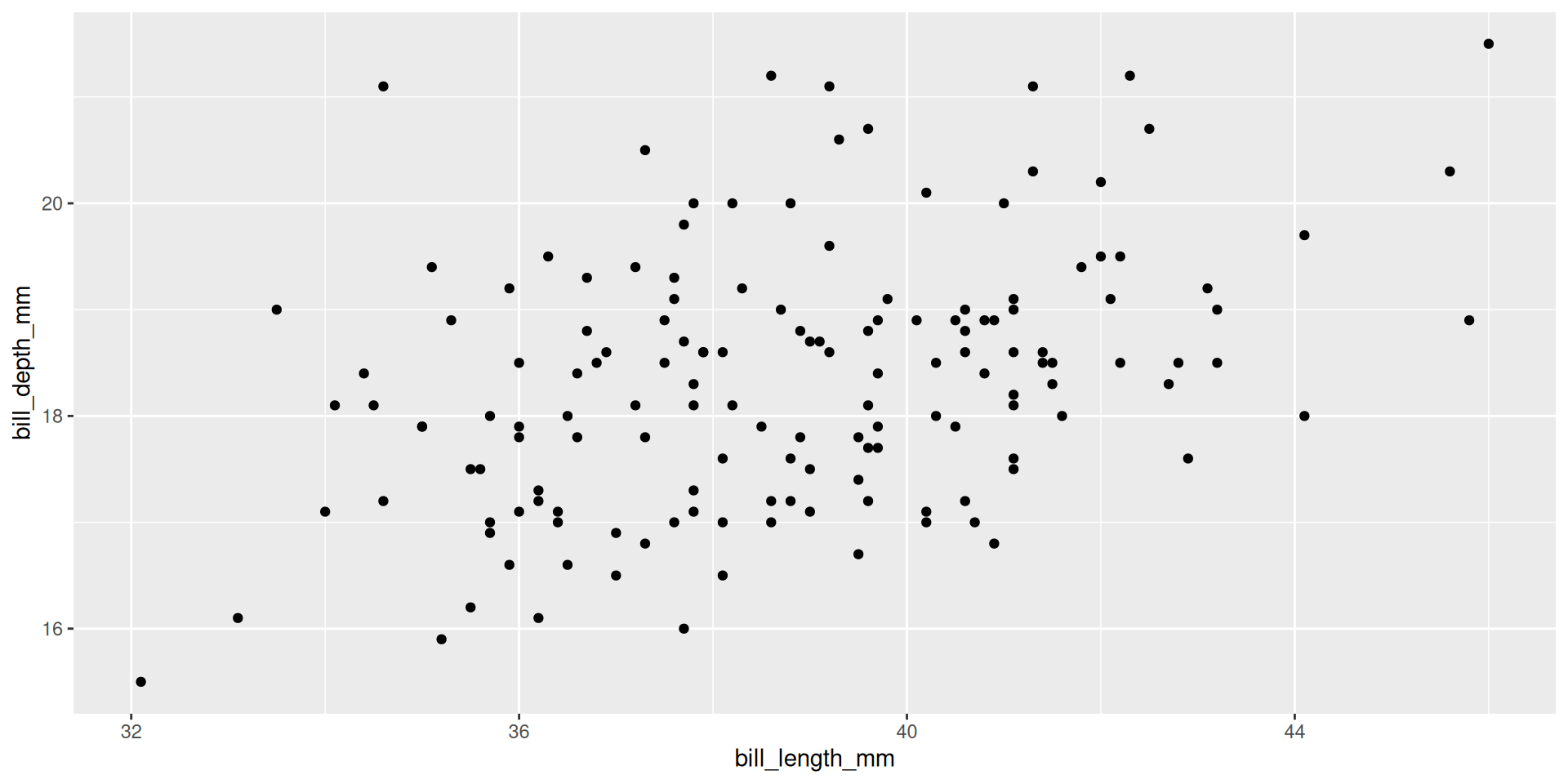
$Chinstrap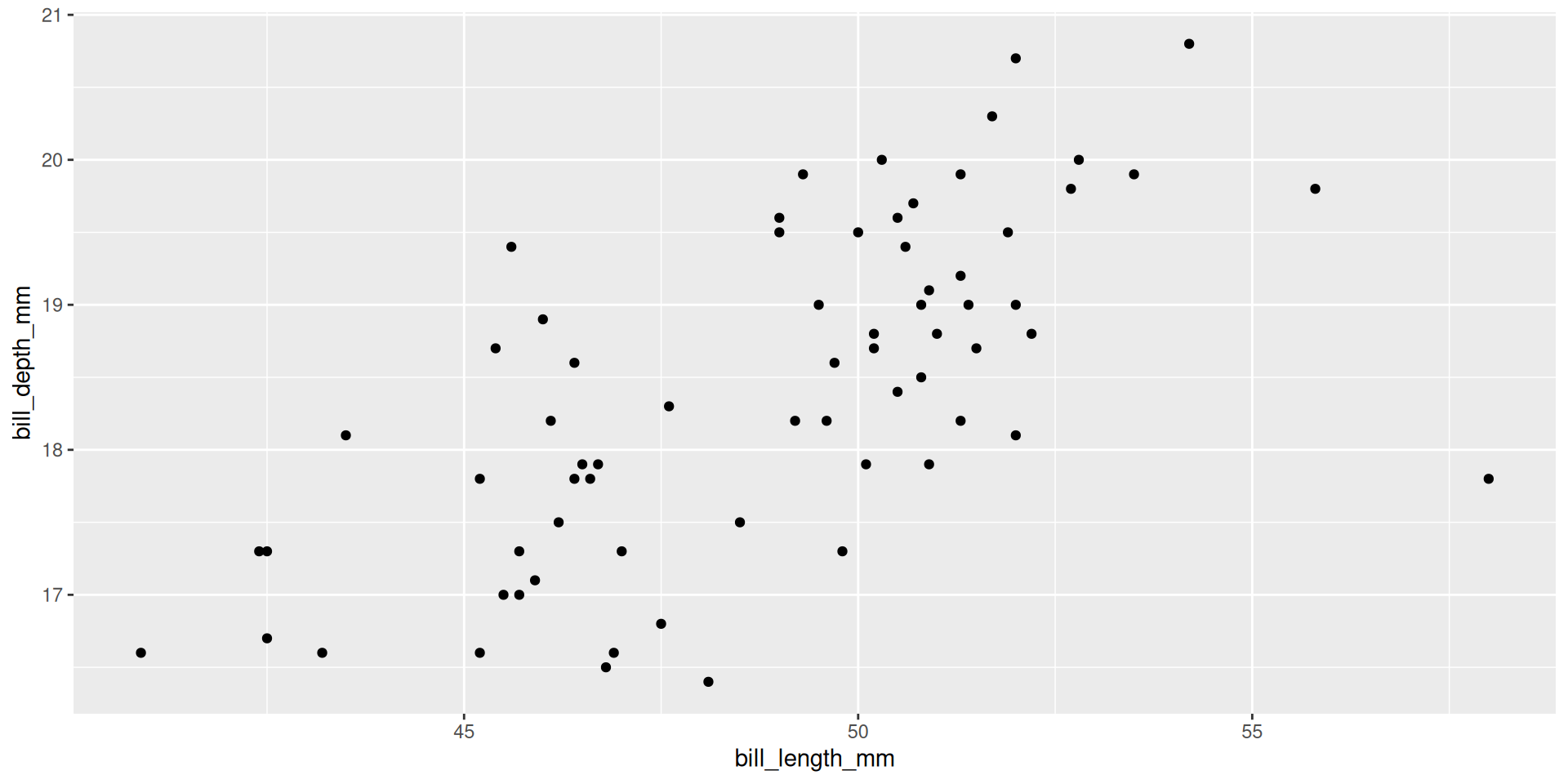
$Gentoo
Importing multiple data files
First, we’ll create multiple data files
species_list <- penguins |>
split(penguins$species)
map2(species_list, names(species_list),
~ write_csv(.x, here(paste0("data/", tolower(.y), "_penguin_data.csv"))))$Adelie
# A tibble: 152 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 142 more rows
# ℹ 2 more variables: sex <fct>, year <int>
$Chinstrap
# A tibble: 68 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Chinstrap Dream 46.5 17.9 192 3500
2 Chinstrap Dream 50 19.5 196 3900
3 Chinstrap Dream 51.3 19.2 193 3650
4 Chinstrap Dream 45.4 18.7 188 3525
5 Chinstrap Dream 52.7 19.8 197 3725
6 Chinstrap Dream 45.2 17.8 198 3950
7 Chinstrap Dream 46.1 18.2 178 3250
8 Chinstrap Dream 51.3 18.2 197 3750
9 Chinstrap Dream 46 18.9 195 4150
10 Chinstrap Dream 51.3 19.9 198 3700
# ℹ 58 more rows
# ℹ 2 more variables: sex <fct>, year <int>
$Gentoo
# A tibble: 124 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Gentoo Biscoe 46.1 13.2 211 4500
2 Gentoo Biscoe 50 16.3 230 5700
3 Gentoo Biscoe 48.7 14.1 210 4450
4 Gentoo Biscoe 50 15.2 218 5700
5 Gentoo Biscoe 47.6 14.5 215 5400
6 Gentoo Biscoe 46.5 13.5 210 4550
7 Gentoo Biscoe 45.4 14.6 211 4800
8 Gentoo Biscoe 46.7 15.3 219 5200
9 Gentoo Biscoe 43.3 13.4 209 4400
10 Gentoo Biscoe 46.8 15.4 215 5150
# ℹ 114 more rows
# ℹ 2 more variables: sex <fct>, year <int>Importing multiple data files
Get file names and paths with dir()
(penguin_files <- dir(path = "../data",
pattern = "penguin_data.csv",
full.names = TRUE))[1] "../data/adelie_penguin_data.csv" "../data/chinstrap_penguin_data.csv"
[3] "../data/gentoo_penguin_data.csv" Importing multiple data files
Map readr::read_csv() to each element of penguin_files
(penguin_data1 <- map(penguin_files, read_csv))[[1]]
# A tibble: 152 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 142 more rows
# ℹ 2 more variables: sex <chr>, year <dbl>
[[2]]
# A tibble: 68 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Chinstrap Dream 46.5 17.9 192 3500
2 Chinstrap Dream 50 19.5 196 3900
3 Chinstrap Dream 51.3 19.2 193 3650
4 Chinstrap Dream 45.4 18.7 188 3525
5 Chinstrap Dream 52.7 19.8 197 3725
6 Chinstrap Dream 45.2 17.8 198 3950
7 Chinstrap Dream 46.1 18.2 178 3250
8 Chinstrap Dream 51.3 18.2 197 3750
9 Chinstrap Dream 46 18.9 195 4150
10 Chinstrap Dream 51.3 19.9 198 3700
# ℹ 58 more rows
# ℹ 2 more variables: sex <chr>, year <dbl>
[[3]]
# A tibble: 124 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Gentoo Biscoe 46.1 13.2 211 4500
2 Gentoo Biscoe 50 16.3 230 5700
3 Gentoo Biscoe 48.7 14.1 210 4450
4 Gentoo Biscoe 50 15.2 218 5700
5 Gentoo Biscoe 47.6 14.5 215 5400
6 Gentoo Biscoe 46.5 13.5 210 4550
7 Gentoo Biscoe 45.4 14.6 211 4800
8 Gentoo Biscoe 46.7 15.3 219 5200
9 Gentoo Biscoe 43.3 13.4 209 4400
10 Gentoo Biscoe 46.8 15.4 215 5150
# ℹ 114 more rows
# ℹ 2 more variables: sex <chr>, year <dbl>Importing multiple data files
Use purrr::list_rbind() to return output as data frame
(penguin_data2 <- map(penguin_files, read_csv) |>
list_rbind())# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 334 more rows
# ℹ 2 more variables: sex <chr>, year <dbl>Solving the problems
- Run the same model on multiple data sets
- Create the same plot for multiple data sets
- Read in data files from multiple subjects
