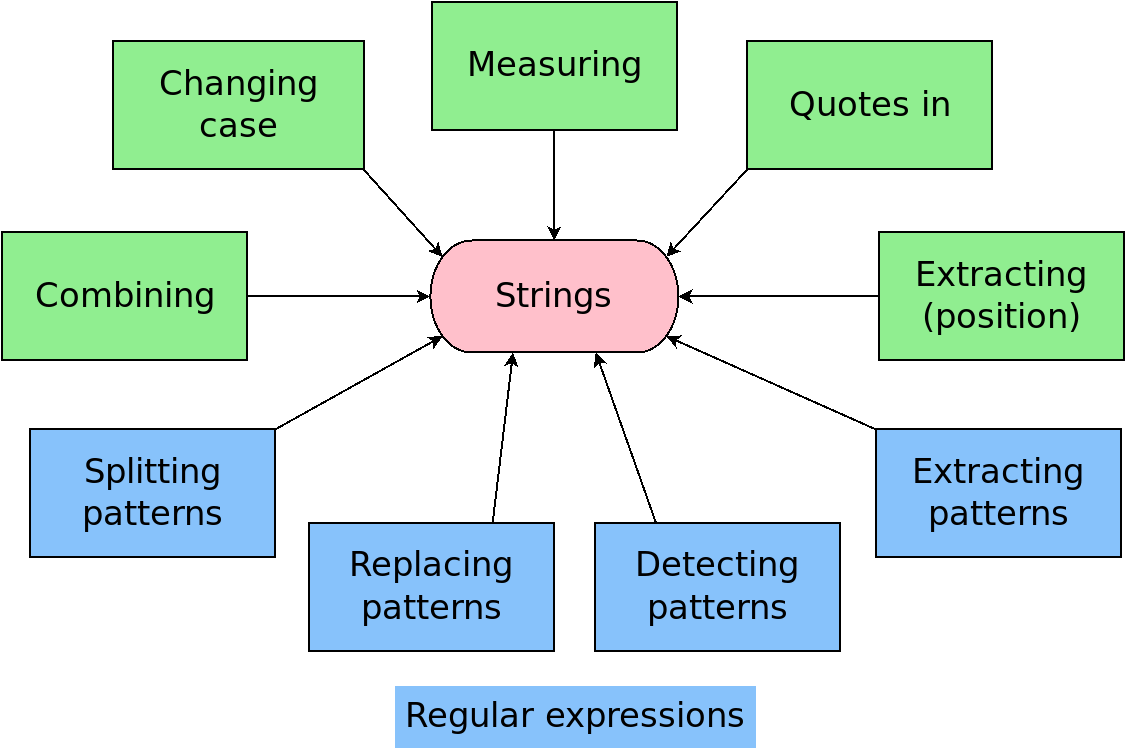
data1# A tibble: 12 × 3
time species resp
<chr> <chr> <chr>
1 early-day dogfish yes
2 mid-day bear dog no
3 late-day dog yes
4 daytime dogfish no
5 early-day cat yes
6 mid-day cat no
7 late-day dogfish no
8 daytime bear dog no
9 early-day dogfish <NA>
10 mid-day catfish yes
11 late-day cat yes
12 daytime bear dog yes